Research
Publications & Scientific Interests.
My research focused on developing new approaches to understanding disease progression, particularly the dynamics of neurodegenerative diseases, using machine learning tools.
The aim was to design better clinical trials tailored specifically for neurodegenerative conditions.
The goals of my research are three-fold:
- Gain a deeper understanding of the spatio-temporal patterns of brain alterations, particularly through the development of numerical models of disease progression based on longitudinal data,
- Characterize individual progression patterns by analyzing the influence of cofactors such as gender, genetics, and socio-demographic information, while predicting individual diagnoses and disease stages up to five years in advance.,
- Identify the optimal time for patients to respond to drugs in clinical trials, which involves detecting the biomarkers most likely to reveal drug effects.
To this end, I have introduced several methodological improvements to existing approaches (see Publications). Most of them are based on a range of tools and software I have developed, in particular:
- Leaspy: LEArning Spatiotemporal Patterns in Python. Enables to estimate the diseae progression for various biomarkers at the average and individual level. Python 3.7.
- Leaspype: a software wrapper around Leaspy, dedicated to forecast disease progression and run statistical analysis to enhance clinical trials with prognostic enrichment (with procova methogology)
- Digital Brain: Web application to vizualise the group-average and invidiaul evolution of the cortical thickness, the PET-FDG, the hippocampus mesh and the cognitive scores, during the course of Alzheimer's Disease. Digital-Brain
Scientific Interests & Skills
Applied Maths
Machine Learning
Deep Learning
CNN
Statistical Learning
Riemannian Geometry
EM-like Algorithms
MCMC methods
Coding
C++ 14
Python
Docker (compose)
FastAPI
Django
Pytorch
VueJS
NodeJS
scikit-learn
Language
French
Ukrainian
English
Main publications
Click on the image (or title for phone-users) to read the abstract

Forecasting individual progression trajectories in Alzheimer’s disease
In Nature Communications. 2023. PDF & Link to publication

Forecasting individual progression trajectories in Huntington disease enables more powered clinical trials
In Scientific Reports. 2022. PDF & Link to publication

AD Course Map charts Alzheimer’s disease progression
In Scientific Reports. 2021. PDF & Link to publication
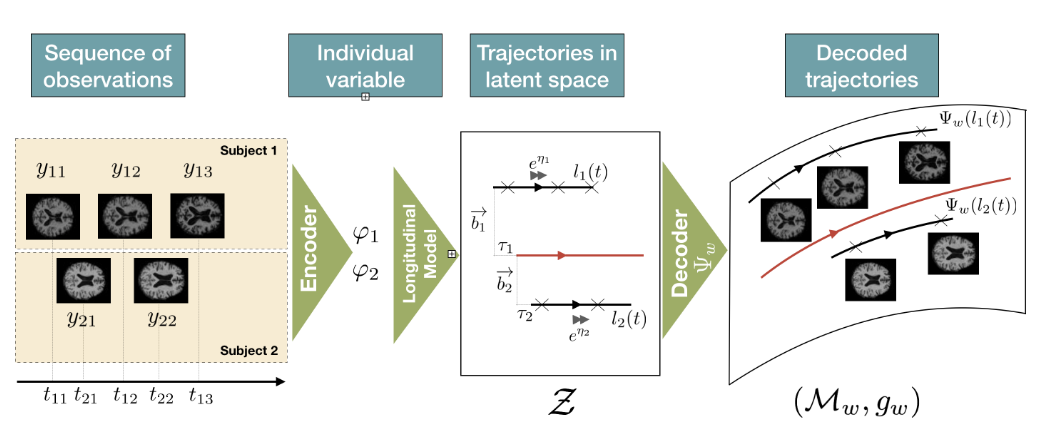
Riemannian Geometry Learning for Disease Progression Modelling
In IPMI. 2019. PDF & Link to publication
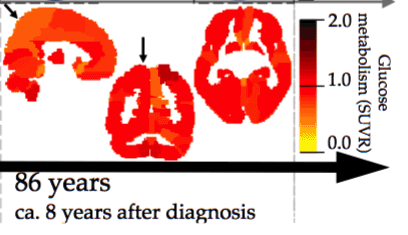
Design of a Decision Support System for Predicting the Progression of Alzheimer's Disease
In Alzheimer's & Dementia. 2018. PDF & Link to publication
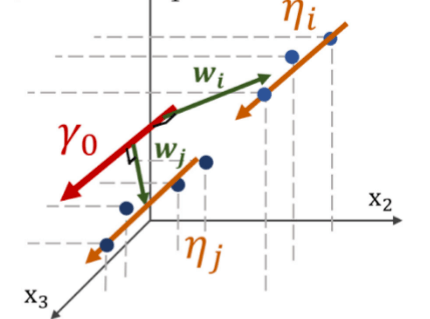
Spatiotemporal Propagation of the Cortical Atrophy: Population and Individual Patterns
In Frontiers in Neurology. 2018. PDF & Link to publication
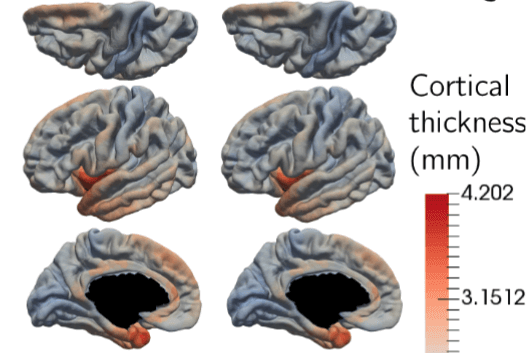
Statistical Learning of Spatiotemporal Patterns from Longitudinal Manifold-Valued Networks
In MICCAI. 2017. PDF & Link to publication
Teaching
Machine Learning in Healthcare @ Data Science Summer School
Summer 2017, 2018 & 2020
Lviv, Ukraine
Courses of Machine Learning, applied to Healthcare, at an international Summer School. They include 5 lectures of 3 hours each, and, a 4 days project, with students (Bachelor, Master and PhD) and workers.
Programming in C
2017 / 2018
University Pierre et Marie Curie
Introductory C classes for licence (Bachelor) students, including practical sessions on computer, blackboard sessions, homeworks and exams
Advanced Machine Learning
2015
Master Data Science, Telecom Paris
Supervision of practical sessions of the Advanced Machine Learniing class of the Data Science master degree
Teachers in charge of the class : Florence d'Alché-Buc & Erwan Le Pennec